| PAnDA requires a template method file that has the instructions for how to analyze your sample. Set up a method
file that fits your needs, then make the following additions/changes: |
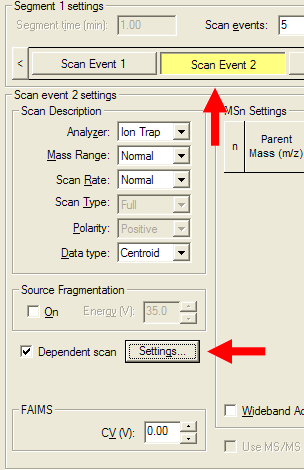
| Select your first MS/MS scan event and bring up the "Settings" dialog. |
 |
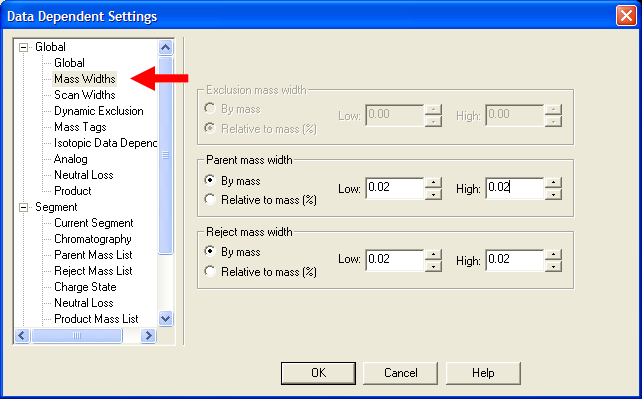
| Under Global->Mass Widths, put in appropriate values. Recommended values are still being evaluated, but putting
mass widths appropriate for high resolution mass spectrometry is a good idea. |
 |
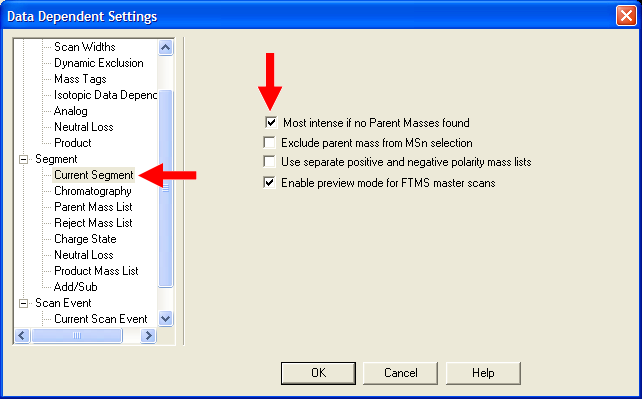
| Under Segment->Current Segment, select "Most intense if no Parent Masses found". This will insure an appropriate
MS/MS event will occur if the mass list does not provide a suggestion. |
 |
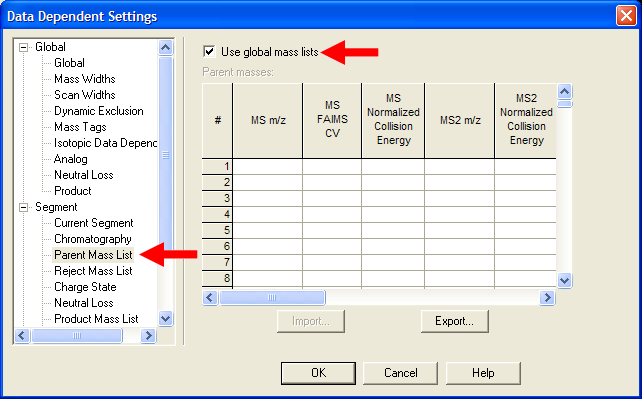
| Under Segment->Parent Mass List, select "Use global mass lists". This setting is critical to using mass lists. |
 |
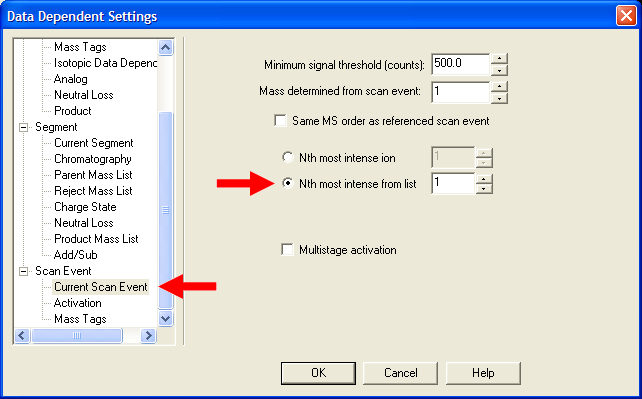
| Under Scan Event->Current Scan Event, select "Nth most intense ion from list". If this is your first MS/MS scan event,
put a value of 1, if this is your second MS/MS scan event, put a value of 2, etc. YOU MUST REPEAT THIS STEP FOR ALL MS/MS SCAN EVENTS!!! |
 |

