Sequence #2, Gene Prediction #1
1. 5
2. No
3. -
4. The exon scores are fairly low, so it is no surprise that the predicted gene doesn't exist.
5. -
Note: a common mistake was to say this prediction matched INS-IGF2. However, the prediction fit entirely
inside an INS-IGF2 intron, thus there was no alignment among exons.
Furthermore, the INS-IGF2 gene is on the complementary strand.
Sequence #2, Gene Prediction #2
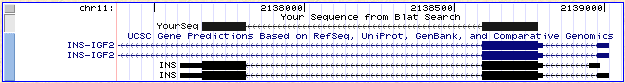
Note: This image is for your reference, it was not required for the homework.
1. 2
2. Yes, INS. There is also partial alignment to INS-IGF2, but only to the exon shared between INS and INS-IGF2
3. 1 exon missed.
4. These exon scores are very confident and the exons do match to exonic sequence in the UCSC browser.
5.
| Exon | INS | GenScan |
| 1 | 38/42 | n/a |
| 2 | 204 | 187 |
| 3 | 219 | 146 |
INS has alternate exon 1 sites. The extra bases in the INS exons are flagged as untranslated in the UCSC browser.
GENSCAN accurately predicts the translated portions of INS.
Sequence #3

Note: This image is for your reference, it was not required for the homework.
1. 6
2. Yes, IL-3
3. 1 exon missed, 2 extra exons predicted.
4. The last two exons in the GENSCAN prediction have low scores. They also do not match any gene sequence.
5.
| Exon | IL-3 | GenScan |
| 1 | 215 | 162 |
| 2 | 42 | 42 |
| 3 | 90 | 90 |
| 4 | 42 | 42 |
| 5a | 535 | n/a |
| 5b | n/a | 51 |
| 6 | n/a | 170 |
The extra bases in the first known exon are flagged as untranslated in the UCSC browser. GENSCAN accurately
predicts the translated portion of the gene up to the 4th exon, but misses the 5th exon and incorrectly predicts
two additional exons.

