Written by Michael J. MacCoss, Department of Genome Sciences, University of Washington
Note: The P3 Predictor's capabilities have been replaced by Skyline. We recommend you check out this new tool if you are performing targeted proteomics experiments using selected reaction monitoring.
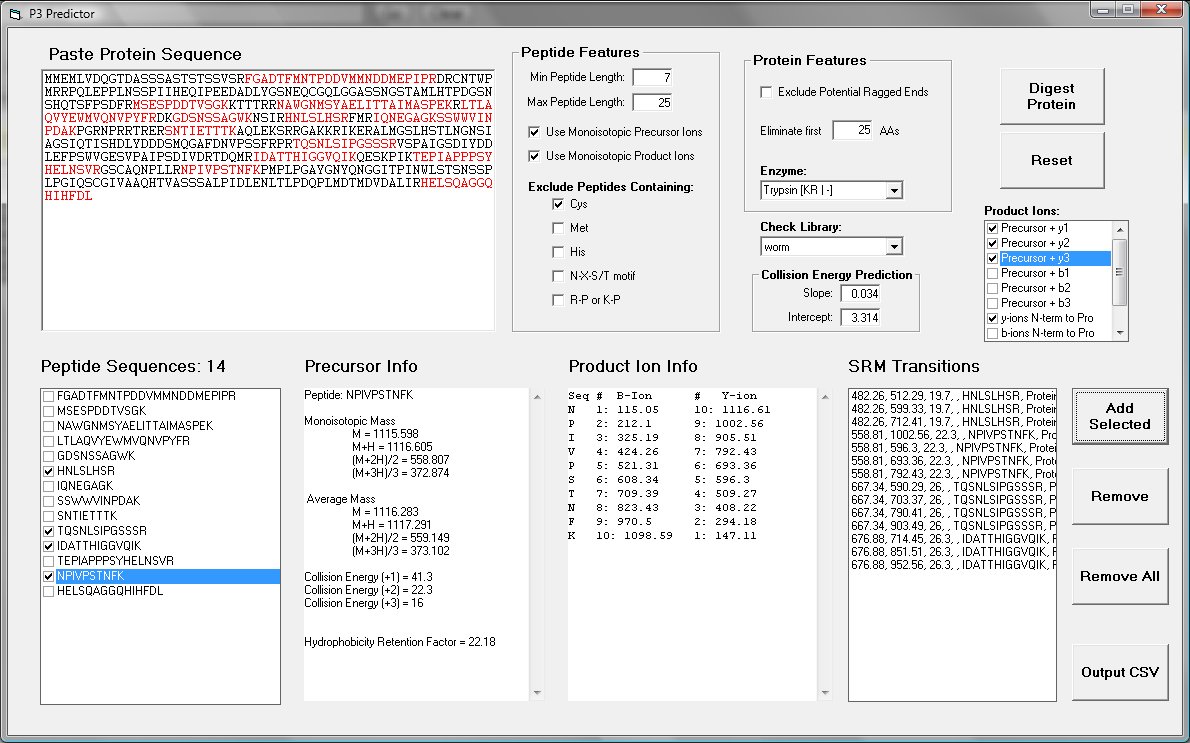
Background: P3 was written to simplify the selection of precursor > product ion transitions for peptides to be measured by selected reaction monitoring.Input: Input is by typing or pasting a protein sequence.
Output: The output file is a comma separated values (CSV) file that can be uploaded into the SRM transition table of Xcalibur for the TSQ Quantum.
Availability: P3 is free for non-profit use. Source code is available upon request. P3 is academic software and its use is at your own risk.
Platform: IDCalc has been tested using Microsoft Windows XP and Vista. Under Vista the user might need to install the latest Visual Basic runtime software. If the software doesn't run under Vista try installing the latest Visual Basic 6.0 runtime.
Installation: Place the P3_Predictor.exe and the associated .stat files in the same directory on your harddrive. The stat files list peptide identified for a given organism in our laboratory. To grab the latest files go to the BiblioSpec library page. The stat files contained in the download zip can be updated with the summary files produced by BiblioSpec.
Download: Right mouse click here and select "Save link as ..."
Note: Cysteines are always assumed to be reduced and alkylated with iodoacetamide. There currently are not any plans to change this in the software.
Screenshot:
P3 is Copyright © 2008 University of Washington. All rights
reserved. Written by Michael J. MacCoss, in the Department of Genome
Sciences at the University
of Washington
MacCoss Lab
Department of Genome Sciences
University of Washington